This is an alternative to plot_boxplot() using base R. This function
should work with most matrix or matrix-like objects (which is especially
useful for DelayedArray objects). The function also includes an
option for computing the relative-log-expression of the input data, given the
input data is on the log-scale.
Usage
plot_boxplot2(
x,
metadata = NULL,
fill_by = NULL,
rle = FALSE,
hcl_palette = "Dark 3",
plot_title = NULL,
x_label = NULL,
y_label = NULL,
show_outliers = TRUE,
show_legend = TRUE,
legend_position = "top",
legend_horiz = TRUE,
legend_cex = 0.8,
legend_ncol = 1,
...
)Arguments
- x
feature x sample matrix or matrix-like object.
- metadata
data.frame containing metadata per sample. rownames of metadata must match the colnames of the input matrix. Default NULL, each sample in the matrix will be plotted.
- fill_by
metadata column used to color boxplots by the grouping variable. Default NULL, each sample in the matrix will be plotted
- rle
should the relative-log-expression value be plotted. Requires input matrix to be on the log-scale. Default = FALSE
- hcl_palette
color palette applied to 'fill_by' variable. One of the
hcl.pals(). Default "Dark 3"- plot_title
title of the plot. Default NULL
- x_label
title of th x-axis. Default NULL
- y_label
title of the y-axis. Default NULL
- show_outliers
should boxplot outliers be shown. Default TRUE
- show_legend
should the legend be drawn on the plot when fill_by is set? Default TRUE
- legend_position
location keyword for the legend. One of "bottomright", "bottom", "bottomleft", "left", "topleft", "top", "topright", "right" and "center". Default "topright"
- legend_horiz
logical; if TRUE, set the legend horizontally rather than vertically. Default TRUE
- legend_cex
character expansion factor for legend text relative to current par. Default 0.8
- legend_ncol
number of columns to set the legend items. Default 1. This is not used if legend_horiz=TRUE
- ...
additional arguments passed to
bxp()
Examples
# Create metadata for plotting
metadata <- data.frame(row.names = colnames(GSE161650_lc))
metadata$Group <- rep(c("DMSO", "THZ1"), each = 3)
# Plot the boxplot by sample
plot_boxplot2(GSE161650_lc)
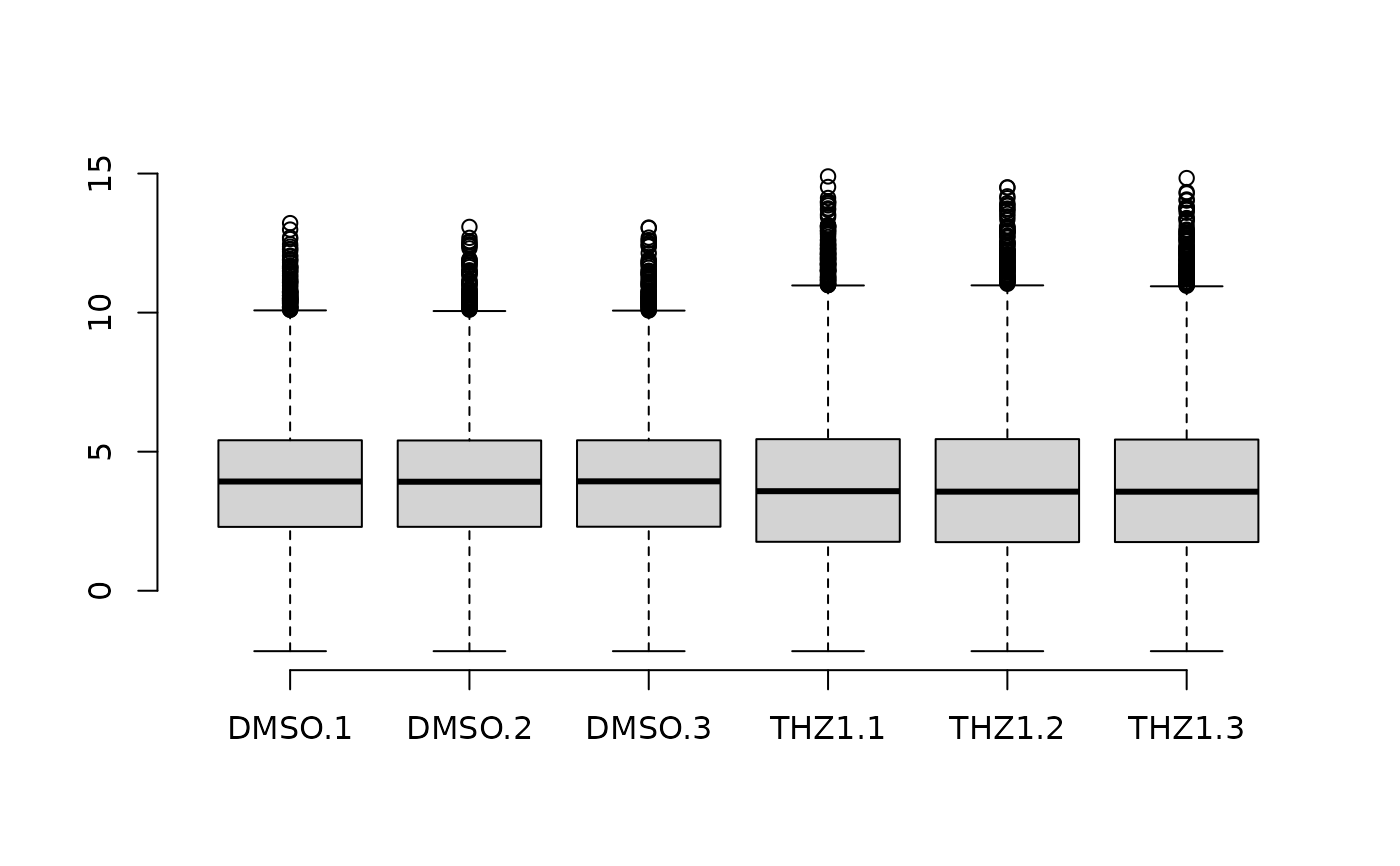 # Plot the boxplot by coloring each Group and transforming values
# to relative-log-expression
plot_boxplot2(
GSE161650_lc,
metadata,
fill_by = "Group",
rle = TRUE,
plot_title = "Relative log-expression",
y_label = "RLE",
show_outliers = FALSE
)
# Plot the boxplot by coloring each Group and transforming values
# to relative-log-expression
plot_boxplot2(
GSE161650_lc,
metadata,
fill_by = "Group",
rle = TRUE,
plot_title = "Relative log-expression",
y_label = "RLE",
show_outliers = FALSE
)