Practical analysis with SummarizedExperiments
Source:vignettes/articles/Practical-analysis-with-SummarizedExperiments.Rmd
Practical-analysis-with-SummarizedExperiments.RmdWhat are SummarizedExperiments
SummarizedExperiments are R objects meant for organizing
and manipulating rectangular matrices that are typically produced by
arrays or high-throughput sequencing. If you are doing any kind of
analysis that requires associating feature-level data (RNA-seq gene
counts, methylation array loci, ATAC-seq regions, etc.) with the genomic
coordinates of those features and the sample-level metadata with which
those features were measured, then you should be using a
SummarizedExperiment to organize, manipulate, and store
your results.
Please take a moment to read through the first 2 sections (at least)
of the SummarizedExperiment
vignette in order to familiarize yourself with what
SummarizedExperiments are and their structure. I will
demonstrate how you can use SummarizedExperiments
below.
Subsetting in R
SummarizedExperiments allow you to quickly and
effectively subset your data in a synchronized fashion that keeps all
sample-level metadata, feature-level matrix data, and genomic
range-level data consistent. The principles of
SummarizedExperiments are derived from base R subsetting
operations. Therefore, in order to become comfortable with
SummarizedExperiments you should be comfortable with some
base R functions.
You may have only ever encountered R from the perspective of the tidyverse. tidyverse
functions provide useful abstractions for munging tidy data
however, most genomics data is often best represented and operated on as
matrices. Keeping your data in matrix format can provide many benefits
as far as speed and code clarity, which in turn helps to ensure
correctness. You can think of matrices as just fancy 2D versions of
vectors. So what are vectors?
Vectors are the main building blocks of most R analyses. Whenever you
use the c() function, like:
x <- c('a', 'b', 'c') you’re creating a vector. You can
do all kinds of cool things with vectors which will prove useful when
working with SummarizedExperiments.
NOTE: the following is heavily inspired by Norm Matloff’s excellent fasteR tutorial. Take a look there to get a brief and concise overview base R. You should also check out the first few chapters of Hadley Wickham’s amazing book Advanced R. The first edition contains some more information on base R.
Subsetting vectors
Below, we’ll use the built-in R constant called LETTERS.
The LETTERS vector is simply a ‘list’ of all uppercase
letters in the Roman alphabet.
LETTERS
#> [1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N" "O" "P" "Q" "R" "S"
#> [20] "T" "U" "V" "W" "X" "Y" "Z"We can subset the vector by position. For example, to get the 3rd
letter we use the [ operator and the position we want to
extract.
LETTERS[3]
#> [1] "C"We can also use a range of positions.
LETTERS[3:7]
#> [1] "C" "D" "E" "F" "G"We don’t have to select sequential elements either. We can extract elements by using another vector of positions.
LETTERS[c(7, 5, 14, 14, 1, 18, 15)]
#> [1] "G" "E" "N" "N" "A" "R" "O"Vectors become really powerful when we start combining them with logical operations.
my_favorite_letters <- c("A", "B", "C")
# See that this produces a logical vector of (TRUE/FALSE) values
# TRUE when LETTERS is one of my_favorite_letters and FALSE otherwise
LETTERS %in% my_favorite_letters
#> [1] TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
#> [25] FALSE FALSE
# We can use that same expression to filter the vector
LETTERS[LETTERS %in% my_favorite_letters]
#> [1] "A" "B" "C"This same kind of subsetting works on vectors that contain numeric data as well. For example, we can filter the measurements of annual flow of water through the Nile river like so:
Nile is another built-in dataset
# Any values strictly greater than 1200
Nile[Nile > 1200]
#> [1] 1210 1230 1370 1210 1250 1260 1220
# Any even number
Nile[Nile %% 2 == 0]
#> [1] 1120 1160 1210 1160 1160 1230 1370 1140 1110 994 1020 960 1180 958 1140
#> [16] 1100 1210 1150 1250 1260 1220 1030 1100 774 840 874 694 940 916 692
#> [31] 1020 1050 726 456 824 702 1120 1100 832 764 768 864 862 698 744
#> [46] 796 1040 944 984 822 1010 676 846 812 742 1040 860 874 848 890
#> [61] 744 838 1050 918 986 1020 906 1170 912 746 718 714 740Subsetting data.frames
But these are just one dimensional vectors. In R we usually deal with data.frames (tibbles for you tidyers) and matrices. Lucky for us, the subsetting operations we learned for vectors work the same way for data.frames and matrices.
Let’s take a look at the built-in ToothGrowth dataset.
The data consists of the length of odontoblasts in 60 guinea pigs
receiving one of three levels of vitamin C by one of two delivery
methods.
head(ToothGrowth)
#> len supp dose
#> 1 4.2 VC 0.5
#> 2 11.5 VC 0.5
#> 3 7.3 VC 0.5
#> 4 5.8 VC 0.5
#> 5 6.4 VC 0.5
#> 6 10.0 VC 0.5The dollar sign $ is used to extract an individual
column from the data.frame, which is just a vector.
head(ToothGrowth$len)
#> [1] 4.2 11.5 7.3 5.8 6.4 10.0We can also use the [[ to get the same thing.
Double-brackets come in handy when your columns are not valid R names
since $ only works when columns are valid names
head(ToothGrowth[["len"]])
#> [1] 4.2 11.5 7.3 5.8 6.4 10.0When subsetting a data.frame in base R the general scheme is:
df[the rows you want, the columns you want]So in order to get the 5th row of the first column we could do:
ToothGrowth[5, 1]
#> [1] 6.4Again, we can combine this kind of thinking to extract rows and columns matching logical conditions. For example, if we want to get all of the animals administered orange juice (‘OJ’)
ToothGrowth[ToothGrowth$supp == "OJ", ]
#> len supp dose
#> 31 15.2 OJ 0.5
#> 32 21.5 OJ 0.5
#> 33 17.6 OJ 0.5
#> 34 9.7 OJ 0.5
#> 35 14.5 OJ 0.5
#> 36 10.0 OJ 0.5
#> 37 8.2 OJ 0.5
#> 38 9.4 OJ 0.5
#> 39 16.5 OJ 0.5
#> 40 9.7 OJ 0.5
#> 41 19.7 OJ 1.0
#> 42 23.3 OJ 1.0
#> 43 23.6 OJ 1.0
#> 44 26.4 OJ 1.0
#> 45 20.0 OJ 1.0
#> 46 25.2 OJ 1.0
#> 47 25.8 OJ 1.0
#> 48 21.2 OJ 1.0
#> 49 14.5 OJ 1.0
#> 50 27.3 OJ 1.0
#> 51 25.5 OJ 2.0
#> 52 26.4 OJ 2.0
#> 53 22.4 OJ 2.0
#> 54 24.5 OJ 2.0
#> 55 24.8 OJ 2.0
#> 56 30.9 OJ 2.0
#> 57 26.4 OJ 2.0
#> 58 27.3 OJ 2.0
#> 59 29.4 OJ 2.0
#> 60 23.0 OJ 2.0We can also combine logical statements. For example, to get all of the rows for animals administered orange juice and with odontoblast length (‘len’) less than 10.
ToothGrowth[ToothGrowth$supp == "OJ" & ToothGrowth$len < 10, ]
#> len supp dose
#> 34 9.7 OJ 0.5
#> 37 8.2 OJ 0.5
#> 38 9.4 OJ 0.5
#> 40 9.7 OJ 0.5
# We can also use the bracket notation to select rows and columns at the same time
ToothGrowth[ToothGrowth$supp == "OJ" & ToothGrowth$len < 10, c("len", "supp")]
#> len supp
#> 34 9.7 OJ
#> 37 8.2 OJ
#> 38 9.4 OJ
#> 40 9.7 OJIt gets annoying typing ToothGrowth every time we want
to subset the data.frame. Base R has a very useful function called
subset() that can help us type less. subset()
essentially ‘looks inside’ the data.frame that you give it for the given
columns and evaluates the expression without having to explicitly tell R
where to find the columns. Think of it like
dplyr::filter().
subset(ToothGrowth, supp == "OJ" & len < 10)
#> len supp dose
#> 34 9.7 OJ 0.5
#> 37 8.2 OJ 0.5
#> 38 9.4 OJ 0.5
#> 40 9.7 OJ 0.5Subsetting matrices
Matrices behave much like data.frames but unlike data.frames matrices can only contain one type of data. This might sound like a limitation at first but you’ll soon come to realize that matrices are very powerful (and fast) to work with in R.
set.seed(123)
# Create some random data that looks like methylation values
(m <- matrix(
data = runif(6 * 10),
ncol = 6,
dimnames = list(
paste0("CpG.", 1:10),
paste0("Sample", 1:6)
)
))
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> CpG.1 0.2875775 0.95683335 0.8895393 0.96302423 0.1428000 0.04583117
#> CpG.2 0.7883051 0.45333416 0.6928034 0.90229905 0.4145463 0.44220007
#> CpG.3 0.4089769 0.67757064 0.6405068 0.69070528 0.4137243 0.79892485
#> CpG.4 0.8830174 0.57263340 0.9942698 0.79546742 0.3688455 0.12189926
#> CpG.5 0.9404673 0.10292468 0.6557058 0.02461368 0.1524447 0.56094798
#> CpG.6 0.0455565 0.89982497 0.7085305 0.47779597 0.1388061 0.20653139
#> CpG.7 0.5281055 0.24608773 0.5440660 0.75845954 0.2330341 0.12753165
#> CpG.8 0.8924190 0.04205953 0.5941420 0.21640794 0.4659625 0.75330786
#> CpG.9 0.5514350 0.32792072 0.2891597 0.31818101 0.2659726 0.89504536
#> CpG.10 0.4566147 0.95450365 0.1471136 0.23162579 0.8578277 0.37446278If we want to extract the value for CpG.3 for Sample3
m[3, 3]
#> [1] 0.6405068Or all values of CpG.3 for every sample
m[3, ]
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> 0.4089769 0.6775706 0.6405068 0.6907053 0.4137243 0.7989248
# Or refer to the row by it's name
m["CpG.3", ]
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> 0.4089769 0.6775706 0.6405068 0.6907053 0.4137243 0.7989248Or all CpGs for Sample3
m[, 3]
#> CpG.1 CpG.2 CpG.3 CpG.4 CpG.5 CpG.6 CpG.7 CpG.8
#> 0.8895393 0.6928034 0.6405068 0.9942698 0.6557058 0.7085305 0.5440660 0.5941420
#> CpG.9 CpG.10
#> 0.2891597 0.1471136
# Or refer to the column by it's name
m[, "Sample3"]
#> CpG.1 CpG.2 CpG.3 CpG.4 CpG.5 CpG.6 CpG.7 CpG.8
#> 0.8895393 0.6928034 0.6405068 0.9942698 0.6557058 0.7085305 0.5440660 0.5941420
#> CpG.9 CpG.10
#> 0.2891597 0.1471136We can also apply a mask to the entire matrix at once. For example,
the following will mark any value that is greater than 0.5 with
TRUE
m > 0.5
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> CpG.1 FALSE TRUE TRUE TRUE FALSE FALSE
#> CpG.2 TRUE FALSE TRUE TRUE FALSE FALSE
#> CpG.3 FALSE TRUE TRUE TRUE FALSE TRUE
#> CpG.4 TRUE TRUE TRUE TRUE FALSE FALSE
#> CpG.5 TRUE FALSE TRUE FALSE FALSE TRUE
#> CpG.6 FALSE TRUE TRUE FALSE FALSE FALSE
#> CpG.7 TRUE FALSE TRUE TRUE FALSE FALSE
#> CpG.8 TRUE FALSE TRUE FALSE FALSE TRUE
#> CpG.9 TRUE FALSE FALSE FALSE FALSE TRUE
#> CpG.10 FALSE TRUE FALSE FALSE TRUE FALSEWe can use this kind of masking to filter rows of the matrix using
some very helpful base R functions that operate on matrices. For
example, to get only those CpGs where 3 or more samples have a value
> 0.5 we can use the rowSums() like so:
m[rowSums(m > 0.5) > 3, ]
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> CpG.3 0.4089769 0.6775706 0.6405068 0.6907053 0.4137243 0.7989248
#> CpG.4 0.8830174 0.5726334 0.9942698 0.7954674 0.3688455 0.1218993This pattern is very common when dealing with sequencing data. Base R
functions like rowSums() and colMeans() are
specialized to operate over matrices and are the most efficient way to
summarize matrix data. The R package matrixStats
also contains highly optimized functions for operating on matrices.
Compare the above to the tidy solution given the same
matrix.
tidyr::as_tibble(m, rownames = "CpG") |>
tidyr::pivot_longer(!CpG, names_to = "SampleName", values_to = "beta") |>
dplyr::group_by(CpG) |>
dplyr::mutate(n = sum(beta > 0.5)) |>
dplyr::filter(n > 3) |>
tidyr::pivot_wider(id_cols = CpG, names_from = "SampleName", values_from = "beta") |>
tibble::column_to_rownames(var = "CpG") |>
data.matrix()
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> CpG.3 0.4089769 0.6775706 0.6405068 0.6907053 0.4137243 0.7989248
#> CpG.4 0.8830174 0.5726334 0.9942698 0.7954674 0.3688455 0.1218993There’s probably some kind of tidy solution using
across() that I’m missing but this is how most of the tidy
code in the wild that I have seen looks
Hopefully these examples are enough to get you started understanding
how subsetting works in R and appreciate how useful it is. Now that you
have some familiarity with the using R functions for subsetting objects,
you’re ready to start working with
SummarizedExperiments.
The SummarizedExperiment
From the SummarizedExperiment vignette:
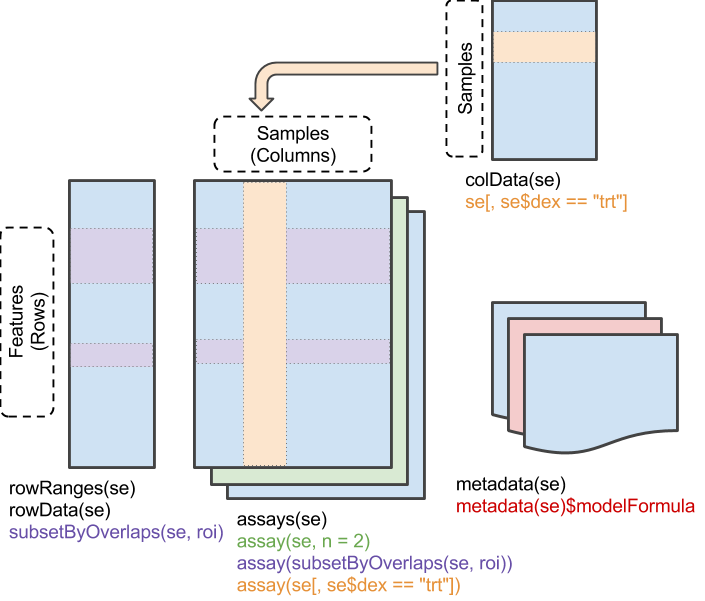
The SummarizedExperiment object coordinates four main
parts:
-
assay(),assays(): A matrix-like or list of matrix-like objects of identical dimension- matrix-like: implements
dim(),dimnames(), and 2-dimensional[,[<-methods. - rows: genes, genomic coordinates, etc.
- columns: samples, cells, etc.
- matrix-like: implements
-
colData(): Annotations on each column, as a DataFrame.- E.g., description of each sample
-
rowData()and / orrowRanges(): Annotations on each row.-
rowRanges(): coordinates of gene / exons in transcripts / etc. -
rowData(): P-values and log-fold change of each gene after differential expression analysis.
-
-
metadata(): List of unstructured metadata describing the overall content of the object.
In order to better understand how they work, let’s construct a
SummarizedExperiment from scratch.
Constructing a SummarizedExperiment
Hopefully you’ll already be working with data that is in a
SummarizedExperiment or some other class that derives from
one. But just in case you don’t have data structured as a
SummarizedExperiment it’s useful and instructive to
understand how to create one from scratch.
To be most useful, a SummarizedExperiment should have at
least:
- A matrix of data with features in rows and samples in columns
- A metadata data.frame with samples as rownames and columns describing their properties
Another really useful object to add to
SummarizedExperiments is a GRanges object describing the
genomic locations of each feature in the matrix. Adding this to the
SummarizedExperiment creates what is called a
RangedSummarizedExperiment that acts just like a regular
SummarizedExperiment with some extra features.
To construct our basic SummarizedExperiment:
- We’ll create a ‘counts’ matrix with gene IDs as rows and Samples in columns
- We’ll add some metadata describing the Samples
- We’ll add on
GRanges()describing the genomic location of the genes
Construct the counts matrix
suppressPackageStartupMessages(library(SummarizedExperiment))
counts <- matrix(
data = rnbinom(n = 200 * 6, mu = 100, size = 1 / 0.5),
nrow = 200,
dimnames = list(paste0("Gene", 1:200), paste0("Sample", 1:6))
)
# Take a peek at what this looks like
counts[1:5, 1:5]
#> Sample1 Sample2 Sample3 Sample4 Sample5
#> Gene1 100 210 186 14 95
#> Gene2 74 17 62 48 27
#> Gene3 129 72 105 203 73
#> Gene4 73 80 84 81 59
#> Gene5 17 242 32 21 58Construct the sample metadata
It is important that the sample metadata be either a
data.frame or a DataFrame object because
SummarizedExperiment requires the colData() to
have rownames that match the colnames of the count matrix.
coldata <- data.frame(
SampleName = colnames(counts),
Treatment = gl(2, 3, labels = c("Control", "Treatment")),
Age = sample.int(100, 6),
row.names = colnames(counts)
)
# Take a peek at what this looks like
coldata
#> SampleName Treatment Age
#> Sample1 Sample1 Control 38
#> Sample2 Sample2 Control 71
#> Sample3 Sample3 Control 30
#> Sample4 Sample4 Treatment 64
#> Sample5 Sample5 Treatment 8
#> Sample6 Sample6 Treatment 47Notice that all of the rownames of the metadata are in the same order as the colnames of the counts matrix. This is necessary.
Construct gene range annotations
You will usually have gene annotations or GRanges
objects loaded from a GTF file or you may even create
GRanges yourself by specifying the chromosome, start, end,
and strand, information manually.
rowranges <- GRanges(
rep(c("chr1", "chr2"), c(50, 150)),
IRanges(floor(runif(200, 1e5, 1e6)), width = 100),
strand = sample(c("+", "-"), 200, TRUE),
feature_id = sprintf("ID%03d", 1:200),
gene_type = sample(c("protein_coding", "lncRNA", "repeat_element"), 200, replace = TRUE)
)
names(rowranges) <- rownames(counts)
# Take a peek at what this looks like
rowranges
#> GRanges object with 200 ranges and 2 metadata columns:
#> seqnames ranges strand | feature_id gene_type
#> <Rle> <IRanges> <Rle> | <character> <character>
#> Gene1 chr1 954187-954286 - | ID001 protein_coding
#> Gene2 chr1 552363-552462 + | ID002 protein_coding
#> Gene3 chr1 303793-303892 - | ID003 lncRNA
#> Gene4 chr1 875980-876079 + | ID004 protein_coding
#> Gene5 chr1 427141-427240 - | ID005 repeat_element
#> ... ... ... ... . ... ...
#> Gene196 chr2 268871-268970 + | ID196 repeat_element
#> Gene197 chr2 599914-600013 + | ID197 repeat_element
#> Gene198 chr2 477464-477563 + | ID198 repeat_element
#> Gene199 chr2 461424-461523 - | ID199 repeat_element
#> Gene200 chr2 619939-620038 + | ID200 protein_coding
#> -------
#> seqinfo: 2 sequences from an unspecified genome; no seqlengthsConstruct the SummarizedExperiment object
With these pieces of information we’re ready to create a
SummarizedExperiment object.
se <- SummarizedExperiment(
assays = list(counts = counts),
colData = coldata,
rowRanges = rowranges
)
# Printing the object gives a summary of what's inside
se
#> class: RangedSummarizedExperiment
#> dim: 200 6
#> metadata(0):
#> assays(1): counts
#> rownames(200): Gene1 Gene2 ... Gene199 Gene200
#> rowData names(2): feature_id gene_type
#> colnames(6): Sample1 Sample2 ... Sample5 Sample6
#> colData names(3): SampleName Treatment AgeAccessing parts of the SummarizedExperiment object
Every part of the SummarizedExperiment object can be
extracted with its accessor function. To extract a particular assay you
can use the assay() function. To extract the column
metadata you can use the colData() function. To extract the
GRanges for the rows of the matrix you can use the
rowRanges() function. The rowData() function
also allows you to access row-level annotation information from data
added to the rowData slot or by the mcols() of
the rowRanges. This will be made more clear below.
Getting the count matrix
assay(se, "counts") |> head()
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> Gene1 100 210 186 14 95 43
#> Gene2 74 17 62 48 27 71
#> Gene3 129 72 105 203 73 63
#> Gene4 73 80 84 81 59 153
#> Gene5 17 242 32 21 58 40
#> Gene6 22 91 38 89 164 43To see what assays are available you can use the
assays() function
assays(se)
#> List of length 1
#> names(1): countsGetting the column metadata
colData(se)
#> DataFrame with 6 rows and 3 columns
#> SampleName Treatment Age
#> <character> <factor> <integer>
#> Sample1 Sample1 Control 38
#> Sample2 Sample2 Control 71
#> Sample3 Sample3 Control 30
#> Sample4 Sample4 Treatment 64
#> Sample5 Sample5 Treatment 8
#> Sample6 Sample6 Treatment 47Getting the rowRanges
rowRanges(se)
#> GRanges object with 200 ranges and 2 metadata columns:
#> seqnames ranges strand | feature_id gene_type
#> <Rle> <IRanges> <Rle> | <character> <character>
#> Gene1 chr1 954187-954286 - | ID001 protein_coding
#> Gene2 chr1 552363-552462 + | ID002 protein_coding
#> Gene3 chr1 303793-303892 - | ID003 lncRNA
#> Gene4 chr1 875980-876079 + | ID004 protein_coding
#> Gene5 chr1 427141-427240 - | ID005 repeat_element
#> ... ... ... ... . ... ...
#> Gene196 chr2 268871-268970 + | ID196 repeat_element
#> Gene197 chr2 599914-600013 + | ID197 repeat_element
#> Gene198 chr2 477464-477563 + | ID198 repeat_element
#> Gene199 chr2 461424-461523 - | ID199 repeat_element
#> Gene200 chr2 619939-620038 + | ID200 protein_coding
#> -------
#> seqinfo: 2 sequences from an unspecified genome; no seqlengthsGetting the rowData
Note that rowData in this case is the same as mcols() of
the rowRanges
rowData(se)
#> DataFrame with 200 rows and 2 columns
#> feature_id gene_type
#> <character> <character>
#> Gene1 ID001 protein_coding
#> Gene2 ID002 protein_coding
#> Gene3 ID003 lncRNA
#> Gene4 ID004 protein_coding
#> Gene5 ID005 repeat_element
#> ... ... ...
#> Gene196 ID196 repeat_element
#> Gene197 ID197 repeat_element
#> Gene198 ID198 repeat_element
#> Gene199 ID199 repeat_element
#> Gene200 ID200 protein_codingModifying a SummarizedExperiment
Once you create a SummarizedExperiment you are not stuck
with the information in that object. SummarizedExperiments
allow you to add and modify the data within the object.
Adding assays
For example, we may wish to calculate counts per million values for
our counts matrix and add a new assay back into our
SummarizedExperiment object.
# Calculate counts per million
counts <- assay(se, "counts")
cpm <- counts / colSums(counts) * 1e6
# Add the CPM data as a new assay to our existing se object
assay(se, "cpm") <- cpm
# And if we wish to log-scale these values
assay(se, "logcounts") <- log2(cpm)
# Now there are three assays available
assays(se)
#> List of length 3
#> names(3): counts cpm logcountsNote: Instead of creating intermediate variables we could also directly use the assays like so:
Adding metadata
SummarizedExperiment objects use the $ to
get and set columns of the metadata contained in the
colData slot. For example, to get all of the Ages we can
use:
se$Age
#> [1] 38 71 30 64 8 47If we want to add a new column we simply create the new column in the same way
se$Batch <- factor(rep(c("A", "B", "C"), 2))
# Now you can se that a new 'Batch` column has been added to the colData
colData(se)
#> DataFrame with 6 rows and 4 columns
#> SampleName Treatment Age Batch
#> <character> <factor> <integer> <factor>
#> Sample1 Sample1 Control 38 A
#> Sample2 Sample2 Control 71 B
#> Sample3 Sample3 Control 30 C
#> Sample4 Sample4 Treatment 64 A
#> Sample5 Sample5 Treatment 8 B
#> Sample6 Sample6 Treatment 47 CAdding rowData
We can also modify the data which describes each feature in the
matrix by adding columns to the rowData. For example, let’s create a new
column called Keep if the gene is a protein_coding
gene.
rowData(se)$Keep <- rowData(se)$gene_type == "protein_coding"
rowData(se)
#> DataFrame with 200 rows and 3 columns
#> feature_id gene_type Keep
#> <character> <character> <logical>
#> Gene1 ID001 protein_coding TRUE
#> Gene2 ID002 protein_coding TRUE
#> Gene3 ID003 lncRNA FALSE
#> Gene4 ID004 protein_coding TRUE
#> Gene5 ID005 repeat_element FALSE
#> ... ... ... ...
#> Gene196 ID196 repeat_element FALSE
#> Gene197 ID197 repeat_element FALSE
#> Gene198 ID198 repeat_element FALSE
#> Gene199 ID199 repeat_element FALSE
#> Gene200 ID200 protein_coding TRUESubsetting SummarizedExperiment objects
SummarizedExperiments follow the basic idea of
se[the rows you want, the columns you want]With a SummarizedExperiment “the rows you want”
corresponds to the features in the rows of the matrix/rowData and “the
columns you want” corresponds to the metadata in
colData
Subsetting based on sample metadata
For example, if we want to select all of the data belonging only to samples in the Treatment group we can use the following:
(trt <- se[, se$Treatment == "Treatment"])
#> class: RangedSummarizedExperiment
#> dim: 200 3
#> metadata(0):
#> assays(3): counts cpm logcounts
#> rownames(200): Gene1 Gene2 ... Gene199 Gene200
#> rowData names(3): feature_id gene_type Keep
#> colnames(3): Sample4 Sample5 Sample6
#> colData names(4): SampleName Treatment Age BatchNotice how the dim of the object changed from 6 to 3.
This is because we have selected only the Samples from the original
SummarizedExperiment object from the treatment group. The
cool thing about SummarizedExperiments is that all of the
assays have also been subsetted to reflect this selection!
Take a look at the “logcounts” assay. It only contains Samples 4, 5, and 6.
assay(trt, "logcounts") |> head()
#> Sample4 Sample5 Sample6
#> Gene1 9.681439 12.23369 11.09368
#> Gene2 11.306469 10.37294 11.82069
#> Gene3 13.329167 11.85724 11.85136
#> Gene4 11.957898 11.55359 12.97889
#> Gene5 10.059737 11.73207 10.98576
#> Gene6 12.146676 13.07906 11.04431Of course you can combine multiple conditions as well
se[, se$Batch %in% c("B", "C") & se$Age > 10]
#> class: RangedSummarizedExperiment
#> dim: 200 3
#> metadata(0):
#> assays(3): counts cpm logcounts
#> rownames(200): Gene1 Gene2 ... Gene199 Gene200
#> rowData names(3): feature_id gene_type Keep
#> colnames(3): Sample2 Sample3 Sample6
#> colData names(4): SampleName Treatment Age BatchSubsetting based on rows
We can also select certain features that we want to keep using row subsetting. For example to select only the first 50 rows:
se[1:50, ]
#> class: RangedSummarizedExperiment
#> dim: 50 6
#> metadata(0):
#> assays(3): counts cpm logcounts
#> rownames(50): Gene1 Gene2 ... Gene49 Gene50
#> rowData names(3): feature_id gene_type Keep
#> colnames(6): Sample1 Sample2 ... Sample5 Sample6
#> colData names(4): SampleName Treatment Age BatchNotice how the dim changed from 200 to 50 reflecting the
fact that we have only selected the first 50 rows.
This subsetting is very useful when combined with logical operators.
Above we created a vector in rowData called “Keep” that
contained TRUE if the corresponding row of the se object was a coding
gene and FALSE otherwise. Let’s use this vector to subset our se
object.
(coding <- se[rowData(se)$Keep, ])
#> class: RangedSummarizedExperiment
#> dim: 68 6
#> metadata(0):
#> assays(3): counts cpm logcounts
#> rownames(68): Gene1 Gene2 ... Gene193 Gene200
#> rowData names(3): feature_id gene_type Keep
#> colnames(6): Sample1 Sample2 ... Sample5 Sample6
#> colData names(4): SampleName Treatment Age BatchAnd if we look at the resulting rowData we can see that
it only contains the protein_coding features
rowData(coding)
#> DataFrame with 68 rows and 3 columns
#> feature_id gene_type Keep
#> <character> <character> <logical>
#> Gene1 ID001 protein_coding TRUE
#> Gene2 ID002 protein_coding TRUE
#> Gene4 ID004 protein_coding TRUE
#> Gene8 ID008 protein_coding TRUE
#> Gene13 ID013 protein_coding TRUE
#> ... ... ... ...
#> Gene183 ID183 protein_coding TRUE
#> Gene185 ID185 protein_coding TRUE
#> Gene187 ID187 protein_coding TRUE
#> Gene193 ID193 protein_coding TRUE
#> Gene200 ID200 protein_coding TRUEEach assay also reflects this operation
assay(coding, "cpm") |> head()
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> Gene1 5865.103 10646.3878 9453.141 821.1144 4816.223 2185.404
#> Gene2 3904.601 834.9295 3158.753 2532.7142 1326.065 3617.281
#> Gene4 3585.286 4075.8101 4432.250 3978.1936 3005.910 8073.027
#> Gene8 1582.946 12180.1483 1069.900 3165.8928 2603.016 815.162
#> Gene13 5571.848 4512.0406 6556.211 12199.4135 9936.629 15704.411
#> Gene15 5019.011 3405.1637 15366.569 6235.7414 4421.630 3988.270Subsetting based on rowRanges
A closely related row-wise subsetting operation can be used if you
have a RangedSummarizedExperiment (a
SummarizedExperiment with a rowRanges slot) that allows you
to perform operations on a SummarizedExperiment object like
you would a GRanges object.
For example, let’s say we only wanted to extract the features on
Chromosome 2 only. Then we can use the GenomicRanges
function subsetByOverlaps directly on our
SummarizedExperiment object like so:
# Region of interest
roi <- GRanges(seqnames = "chr2", ranges = 1:1e7)
# Subset the SE object for only features on chr2
(chr2 <- subsetByOverlaps(se, roi))
#> class: RangedSummarizedExperiment
#> dim: 150 6
#> metadata(0):
#> assays(3): counts cpm logcounts
#> rownames(150): Gene51 Gene52 ... Gene199 Gene200
#> rowData names(3): feature_id gene_type Keep
#> colnames(6): Sample1 Sample2 ... Sample5 Sample6
#> colData names(4): SampleName Treatment Age BatchYou can see again that the dim changed reflecting our
selection. Again, all of the associated assays and rowData have also
been subsetted reflecting this change as well.
rowData(chr2)
#> DataFrame with 150 rows and 3 columns
#> feature_id gene_type Keep
#> <character> <character> <logical>
#> Gene51 ID051 protein_coding TRUE
#> Gene52 ID052 lncRNA FALSE
#> Gene53 ID053 repeat_element FALSE
#> Gene54 ID054 repeat_element FALSE
#> Gene55 ID055 repeat_element FALSE
#> ... ... ... ...
#> Gene196 ID196 repeat_element FALSE
#> Gene197 ID197 repeat_element FALSE
#> Gene198 ID198 repeat_element FALSE
#> Gene199 ID199 repeat_element FALSE
#> Gene200 ID200 protein_coding TRUE
assay(chr2, "counts") |> head()
#> Sample1 Sample2 Sample3 Sample4 Sample5 Sample6
#> Gene51 32 45 139 261 153 95
#> Gene52 113 80 80 61 72 131
#> Gene53 118 24 60 153 5 103
#> Gene54 20 1 35 4 89 49
#> Gene55 249 58 43 307 51 33
#> Gene56 215 96 28 198 50 81
rowRanges(chr2)
#> GRanges object with 150 ranges and 3 metadata columns:
#> seqnames ranges strand | feature_id gene_type Keep
#> <Rle> <IRanges> <Rle> | <character> <character> <logical>
#> Gene51 chr2 839636-839735 + | ID051 protein_coding TRUE
#> Gene52 chr2 388487-388586 + | ID052 lncRNA FALSE
#> Gene53 chr2 357210-357309 + | ID053 repeat_element FALSE
#> Gene54 chr2 230565-230664 + | ID054 repeat_element FALSE
#> Gene55 chr2 491399-491498 + | ID055 repeat_element FALSE
#> ... ... ... ... . ... ... ...
#> Gene196 chr2 268871-268970 + | ID196 repeat_element FALSE
#> Gene197 chr2 599914-600013 + | ID197 repeat_element FALSE
#> Gene198 chr2 477464-477563 + | ID198 repeat_element FALSE
#> Gene199 chr2 461424-461523 - | ID199 repeat_element FALSE
#> Gene200 chr2 619939-620038 + | ID200 protein_coding TRUE
#> -------
#> seqinfo: 2 sequences from an unspecified genome; no seqlengthsThere’s also a few shortcuts on range operations using
GRanges/SummarizedExperiments. See the help pages for
%over, %within%, and %outside%.
For example:
all.equal(se[se %over% roi, ], subsetByOverlaps(se, roi))
#> [1] TRUECombining subsetting operations
Of course you don’t have to perform one subsetting operation at a
time. Like base R you can combine multiple expressions to subset a
SummarizedExperiment object.
For example, to select only features labeled as repeat_elements and the Sample from ‘Batch’ A in the ‘Control’ group
(selected <- se[
rowData(se)$gene_type == "repeat_element",
se$Treatment == "Control" &
se$Batch == "A"
])
#> class: RangedSummarizedExperiment
#> dim: 72 1
#> metadata(0):
#> assays(3): counts cpm logcounts
#> rownames(72): Gene5 Gene6 ... Gene198 Gene199
#> rowData names(3): feature_id gene_type Keep
#> colnames(1): Sample1
#> colData names(4): SampleName Treatment Age BatchSaving a SummarizedExperiment
Since SummarizedExperiments keep basically all
information about an experiment in one place, it is convenient to save
the entire SummarizedExperiment object so that you can pick
up an analysis where you left off or even to facilitate better sharing
of data between collaborators.
You can save the entire SummarizedExperiment object
with:
saveRDS(se, "/path/to/se.rds")
And when you want to read the same object back into R for your next analysis you can do so with:
se <- readRDS("/path/to/se.rds")
SummarizedExperiments in the real world
If you’re working with any Bioconductor packages it’s likely
that the object you’re working with either is a
SummarizedExperiment or is inherited from one. For example,
the DESeqDataSet from the DESeq2
package and BSseq objects from the bsseq
package both inherit from a SummarizedExperiment and
thus retain all of the same functionality as above. If you go to the SummarizedExperiment
landing page and click “See More” under details you can see all of the
packages that depend on SummarizedExperiment.
Also, many common methods are also implemented for
SummarizedExperiment objects. For example, to simplify
calculating counts-per-million above I could have simply used the
edgeR::cpm() directly on the
SummarizedExperiment object. Many functions in bioconductor
packages know how to deal directly with
SummarizedExperiments so you don’t ever have to take the
trouble extracting components and performing tedious calculations
yourself.
I also left out any discussion of the metadata() slot of
the SummarizedExperiment. The metadata slot is simply a
list of any R object that contains information about the experiment. The
metadata in the metadata slots are not subjected to the
same subsetting rules as the other slots. In practice this assay
contains additional information about the experiment as a whole. For
example, I typically store bootstrap alignments for each sample
here.
To add something to the SummarizedExperiment metadata
slot you can do:
metadata(se)$additional_info <- "Experiment performed on 6 samples with three replicates each"And to retrieve this:
metadata(se)$additional_info
#> [1] "Experiment performed on 6 samples with three replicates each"Closing thoughts
Hopefully this was enough information to get you started using
SummarizedExperiments. There’s many things I left out such
as different backings
for storing out of memory data, a tidyverse interface
to SummarizedExperiment objects, TreeSummarizedExperiments
for microbiome data, MultiAssayExperiments
for dealing with experiments containing multiomics data, and much
more.
Please let me know your thoughts and if anything needs more clarification.